Periodic Table |
 |
 |
 |
 |
 |
 |
 |
Lewis Acid/Base Interaction Matrix Database
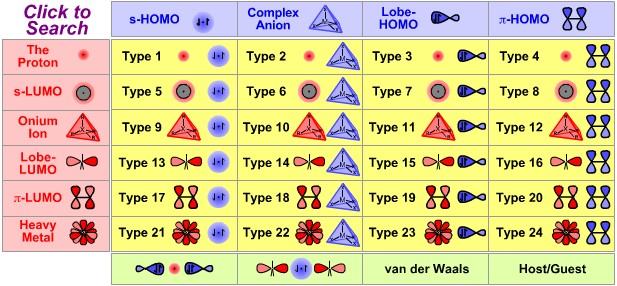 |
Guest-Host Complexation |
|
| Molecular Shape Recognition Complex | |
 |
|
| Bonding: | Guest-Host Complexation "Complexes composed of two or more molecules or ions held together in unique structural relationships by hydrogen bonding, ion pairing or by van der Waals force, but not by full covalent bonds." Wikipedia, p-xylylenediammonium bound within a cucurbituril: 
Molecular Recognition "The term molecular recognition refers to the specific interaction between two or more molecules through non covalent bonding such as hydrogen bonding, metal coordination, hydrophobic forces, van der Waals forces, pi-pi interactions, electrostatic and/or electromagnetic effects. The host and guest involved in molecular recognition exhibit molecular complementarity." Wikipedia
Inclusion Compounds "An inclusion compound is a complexing which one chemical compound, the host, forms a cavity in which molecules of a second guest compound are located. The definition of inclusion compounds is very broad, extending to channels formed between molecules in a crystal lattice in which guest molecules can fit. If the spaces in the host lattice are enclosed on all sides so that the guest species is ‘trapped’ as in a cage, the compound is known as a clathrate. In molecular encapsulation a guest molecule is actually trapped inside another molecule." Wikipedia Solid urea, (NH2)2C=O, can accommodate octane or 1-bromoctane, but not 2-methyl heptane or 2-bromooctane, as guest molecules in a hexagonal host lattice structure which contains long guest channels about 500pm in diameter. Zeolites are aluminosilicate minerals (natural and synthetic) which have open structures able to accommodate a wide range of guest molecules. The microporous molecular structure of a zeolite ZSM-5: 
Biological Systems Molecular recognition plays an important role in biological systems and is observed in between receptor-ligand, antigen-antibody, DNA-protein, sugar-lectin, RNA-ribosome, etc. An example of molecular recognition is the antibiotic vancomycin that selectively binds with the peptides with terminal D-alanyl-D-alanine in bacterial cells through five hydrogen bonds. The vancomycin is lethal to the bacteria since once it has bound to these particular peptides they are unable to be used to construct cell wall. 
Another, out of countless possible examples: Zinc finger proteins coordinate one or more zinc ions to help stabilize their folds. The diagram represents the Cys2His2 zinc finger motif, consisting of an α-helix and an antiparallel β-sheet. The zinc ion (green) is coordinated by two histidine residues and two cysteine residues. 
Zinc finger proteins typically bind DNA, RNA, proteins or small molecules. The diagram represents the protein Zif268 (blue) containing three zinc fingers in complex with DNA (orange). The coordinating amino acid residues and zinc ions (green) are highlighted.  |
| Charge: | Complexes may be negatively charged, positively charged or they may be neutral. |
| Chemistry: | All biochemistry and molecular biology, totally outside the scope of this webbook! |
| Congeneric Series: | See above. |
| van der Waals attraction complex (generic) |
|
|
|
Argon, liquid more here |
|
|
Methyl iodide dimer more here |
|
|
Methyl iodide/Argon complex more here |